2. DeePMD-kit Quick Start Tutorial
DeePMD-kit is a deep learning package for many-body potential energy representation and molecular dynamics.
This tutorial can be directly run on Bohrium Notebook, you can click the
Open in Bohriumbutton above to quickly run this document in Bohrium.After opening Bohrium Notebook, click the button
connect, and choosedeepmd-kit:2.2.1-cuda11.6-notebookas image andc4_m8_cpuas computing resources. Wait a minute and you can get started.
2.1. Task
Mastering the paradigm cycle of using DeePMD-kit to establish deep potential molecular dynamics models, and following a complete case to learn how to apply it to molecular dynamics tasks.
By the end of this tutorial, you will be able to:
Prepare the formataive dataset and running scripts for training with DeePMD-kit;
Train, freeze, and test DeePMD-kit models;
Use DeePMD-kit in LAMMPS for calculations;
Work through this tutorial. It will take you 20 minutes, max!
2.2. Table of contents
Get tutorial data via git
General Introduction
Data preparation
Prepare input script
Train a model
Freeze a model
Test a model
Run MD with LAMMPS
2.3. Get tutorial data via git
! if ! [ -e colombo-academy-tutorials ];then git clone https://gitee.com/deepmodeling/colombo-academy-tutorials.git;fi;
Cloning into 'colombo-academy-tutorials'...
remote: Enumerating objects: 7164, done.
remote: Counting objects: 100% (174/174), done.
remote: Compressing objects: 100% (138/138), done.
remote: Total 7164 (delta 78), reused 71 (delta 32), pack-reused 6990
Receiving objects: 100% (7164/7164), 45.31 MiB | 3.85 MiB/s, done.
Resolving deltas: 100% (3378/3378), done.
Updating files: 100% (185/185), done.
2.4. General Introduction
This tutorial will introduce you to the basic usage of the DeePMD-kit, taking a gas phase methane molecule as an example. DeePMD-kit’s documentation is recommended as the complete reference.
The DP model is generated using the DeePMD-kit package (v2.1.5). The training data is converted into the format of DeePMD-kit using a tool named dpdata (v0.2.14).
Details of dpdata can be found in the dpdata documentation.
We’ve prepared initial data for \(CH_4\) for you, and put them in the folder colombo-academy-tutorials/DeePMD-kit/00.data
import os
prefix_path = os.getcwd()
Folder abacus_md is obtained by performing ab-initio molecular dynamics with ABACUS. Detailed instructions on ABACUS can be found in its document.
os.chdir(
os.path.join(prefix_path, "colombo-academy-tutorials", "DeePMD-kit", "00.data")
)
os.listdir("abacus_md")
['C_ONCV_PBE-1.2.upf',
'C_gga_6au_100Ry_2s2p1d.orb',
'H_ONCV_PBE-1.2.upf',
'H_gga_6au_100Ry_2s1p.orb',
'INPUT',
'KPT',
'OUT.ABACUS',
'STRU']
2.5. Data preparation
The training data utilized by DeePMD-kit comprises essential information such as atom type, simulation box, atom coordinate, atom force, system energy, and virial. A snapshot of a molecular system that includes this data is called a frame. Multiple frames with the same number of atoms and atom types make up a system of data. For instance, a molecular dynamics trajectory can be converted into a system of data, with each time step corresponding to a frame in the system.
To simplify the process of converting data generated by popular simulation software like CP2K, Gaussian, Quantum-Espresso, ABACUS, and LAMMPS into the compressed format of DeePMD-kit, we offer a convenient tool called dpdata.
Next, the data from AIMD is splited randomly as training and validation data.
import dpdata
import numpy as np
# load data of abacus/md format
data = dpdata.LabeledSystem("abacus_md", fmt="abacus/md")
print("# the data contains %d frames" % len(data))
# random choose 40 index for validation_data
rng = np.random.default_rng()
index_validation = rng.choice(201, size=40, replace=False)
# other indexes are training_data
index_training = list(set(range(201)) - set(index_validation))
data_training = data.sub_system(index_training)
data_validation = data.sub_system(index_validation)
# all training data put into directory:"training_data"
data_training.to_deepmd_npy("training_data")
# all validation data put into directory:"validation_data"
data_validation.to_deepmd_npy("validation_data")
print("# the training data contains %d frames" % len(data_training))
print("# the validation data contains %d frames" % len(data_validation))
# the data contains 201 frames
# the training data contains 161 frames
# the validation data contains 40 frames
As you can see, 161 frames are picked as training data, and the other 40 frames are validation dat.
The DeePMD-kit adopts a compressed data format. All training data should first be converted into this format and can then be used by DeePMD-kit. The data format is explained in detail in the DeePMD-kit manual that can be found in the DeePMD-kit Data Introduction.
! tree training_data
training_data
├── set.000
│ ├── box.npy
│ ├── coord.npy
│ ├── energy.npy
│ ├── force.npy
│ └── virial.npy
├── type.raw
└── type_map.raw
1 directory, 7 files
Let’s have a look at type.raw:
! cat training_data/type.raw
0
0
0
0
1
This tells us there are 5 atoms in this example, 4 atoms represented by type “0”, and 1 atom represented by type “1”. Sometimes one needs to map the integer types to atom name. The mapping can be given by the file type_map.raw
! cat training_data/type_map.raw
H
C
This tells us the type “0” is named by “H”, and the type “1” is named by “C”.
More detailed doc about Data conversion can be found here.
2.6. Prepare input script
Once the data preparation is done, we can go on with training. Now go to the training directory
os.chdir(
os.path.join(prefix_path, "colombo-academy-tutorials", "DeePMD-kit", "01.train")
)
DeePMD-kit requires a json format file to specify parameters for training.
In the model section, the parameters of embedding and fitting networks are specified.
"model":{
"type_map": ["H", "C"],
"descriptor":{
"type": "se_e2_a",
"rcut": 6.00,
"rcut_smth": 0.50,
"sel": "auto",
"neuron": [25, 50, 100],
"resnet_dt": false,
"axis_neuron": 16,
"seed": 1,
"_comment": "that's all"
},
"fitting_net":{
"neuron": [240, 240, 240],
"resnet_dt": true,
"seed": 1,
"_comment": "that's all"
},
"_comment": "that's all"'
},
The explanation for some of the parameters is as follows:
Parameter | Expiation |
|---|---|
type_map | the name of each type of atom |
descriptor > type | the type of descriptor |
descriptor > rcut | cut-off radius |
descriptor > rcut_smth | where the smoothing starts |
descriptor > sel | the maximum number of type i atoms in the cut-off radius |
descriptor > neuron | size of the embedding neural network |
descriptor > axis_neuron | the size of the submatrix of G (embedding matrix) |
fitting_net > neuron | size of the fitting neural network |
The se_e2_a descriptor is used to train the DP model. The item neurons set the size of the descriptors and fitting network to [25, 50, 100] and [240, 240, 240], respectively. The components in local environment to smoothly go to zero from 0.5 to 6 Å.
The following are the parameters that specify the learning rate and loss function.
"learning_rate" :{
"type": "exp",
"decay_steps": 50,
"start_lr": 0.001,
"stop_lr": 3.51e-8,
"_comment": "that's all"
},
"loss" :{
"type": "ener",
"start_pref_e": 0.02,
"limit_pref_e": 1,
"start_pref_f": 1000,
"limit_pref_f": 1,
"start_pref_v": 0,
"limit_pref_v": 0,
"_comment": "that's all"
},
In the loss function, pref_e increases from 0.02 to 1, and pref_f decreases from 1000 to 1 progressively, which means that the force term dominates at the beginning, while energy and virial terms become important at the end. This strategy is very effective and reduces the total training time. pref_v is set to 0 , indicating that no virial data are included in the training process. The starting learning rate, stop learning rate, and decay steps are set to 0.001, 3.51e-8, and 50, respectively. The model is trained for 10000 steps.
The training parameters are given in the following
"training" : {
"training_data": {
"systems": ["../00.data/training_data"],
"batch_size": "auto",
"_comment": "that's all"
},
"validation_data":{
"systems": ["../00.data/validation_data/"],
"batch_size": "auto",
"numb_btch": 1,
"_comment": "that's all"
},
"numb_steps": 10000,
"seed": 10,
"disp_file": "lcurve.out",
"disp_freq": 200,
"save_freq": 10000,
},
More detailed docs about Data conversion can be found here.
2.7. Train a model
After the training script is prepared, we can start the training with DeePMD-kit by simply running
! dp train input.json
WARNING:tensorflow:From /opt/deepmd-kit-2.2.1/lib/python3.10/site-packages/tensorflow/python/compat/v2_compat.py:107: disable_resource_variables (from tensorflow.python.ops.variable_scope) is deprecated and will be removed in a future version.
Instructions for updating:
non-resource variables are not supported in the long term
WARNING:root:To get the best performance, it is recommended to adjust the number of threads by setting the environment variables OMP_NUM_THREADS, TF_INTRA_OP_PARALLELISM_THREADS, and TF_INTER_OP_PARALLELISM_THREADS. See https://deepmd.rtfd.io/parallelism/ for more information.
WARNING:root:Environment variable KMP_BLOCKTIME is empty. Use the default value 0
WARNING:root:Environment variable KMP_AFFINITY is empty. Use the default value granularity=fine,verbose,compact,1,0
/opt/deepmd-kit-2.2.1/lib/python3.10/importlib/__init__.py:169: UserWarning: The NumPy module was reloaded (imported a second time). This can in some cases result in small but subtle issues and is discouraged.
_bootstrap._exec(spec, module)
DEEPMD INFO Calculate neighbor statistics... (add --skip-neighbor-stat to skip this step)
2023-04-20 23:35:59.335932: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /usr/local/nvidia/lib:/usr/local/nvidia/lib64
2023-04-20 23:35:59.335979: W tensorflow/stream_executor/cuda/cuda_driver.cc:269] failed call to cuInit: UNKNOWN ERROR (303)
OMP: Info #155: KMP_AFFINITY: Initial OS proc set respected: 0-7
OMP: Info #216: KMP_AFFINITY: decoding x2APIC ids.
OMP: Info #216: KMP_AFFINITY: cpuid leaf 11 not supported.
OMP: Info #216: KMP_AFFINITY: decoding legacy APIC ids.
OMP: Info #157: KMP_AFFINITY: 8 available OS procs
OMP: Info #158: KMP_AFFINITY: Uniform topology
OMP: Info #287: KMP_AFFINITY: topology layer "LL cache" is equivalent to "socket".
OMP: Info #192: KMP_AFFINITY: 1 socket x 4 cores/socket x 2 threads/core (4 total cores)
OMP: Info #218: KMP_AFFINITY: OS proc to physical thread map:
OMP: Info #172: KMP_AFFINITY: OS proc 0 maps to socket 0 core 0 thread 0
OMP: Info #172: KMP_AFFINITY: OS proc 1 maps to socket 0 core 0 thread 1
OMP: Info #172: KMP_AFFINITY: OS proc 2 maps to socket 0 core 1 thread 0
OMP: Info #172: KMP_AFFINITY: OS proc 3 maps to socket 0 core 1 thread 1
OMP: Info #172: KMP_AFFINITY: OS proc 4 maps to socket 0 core 2 thread 0
OMP: Info #172: KMP_AFFINITY: OS proc 5 maps to socket 0 core 2 thread 1
OMP: Info #172: KMP_AFFINITY: OS proc 6 maps to socket 0 core 3 thread 0
OMP: Info #172: KMP_AFFINITY: OS proc 7 maps to socket 0 core 3 thread 1
OMP: Info #254: KMP_AFFINITY: pid 118 tid 140 thread 1 bound to OS proc set 2
OMP: Info #254: KMP_AFFINITY: pid 118 tid 142 thread 2 bound to OS proc set 4
OMP: Info #254: KMP_AFFINITY: pid 118 tid 144 thread 4 bound to OS proc set 1
OMP: Info #254: KMP_AFFINITY: pid 118 tid 143 thread 3 bound to OS proc set 6
OMP: Info #254: KMP_AFFINITY: pid 118 tid 145 thread 5 bound to OS proc set 3
OMP: Info #254: KMP_AFFINITY: pid 118 tid 146 thread 6 bound to OS proc set 5
OMP: Info #254: KMP_AFFINITY: pid 118 tid 147 thread 7 bound to OS proc set 7
OMP: Info #254: KMP_AFFINITY: pid 118 tid 148 thread 8 bound to OS proc set 0
OMP: Info #254: KMP_AFFINITY: pid 118 tid 139 thread 9 bound to OS proc set 2
OMP: Info #254: KMP_AFFINITY: pid 118 tid 149 thread 10 bound to OS proc set 4
OMP: Info #254: KMP_AFFINITY: pid 118 tid 150 thread 11 bound to OS proc set 6
OMP: Info #254: KMP_AFFINITY: pid 118 tid 151 thread 12 bound to OS proc set 1
OMP: Info #254: KMP_AFFINITY: pid 118 tid 152 thread 13 bound to OS proc set 3
OMP: Info #254: KMP_AFFINITY: pid 118 tid 153 thread 14 bound to OS proc set 5
OMP: Info #254: KMP_AFFINITY: pid 118 tid 154 thread 15 bound to OS proc set 7
OMP: Info #254: KMP_AFFINITY: pid 118 tid 155 thread 16 bound to OS proc set 0
DEEPMD INFO training data with min nbor dist: 1.045920568611028
DEEPMD INFO training data with max nbor size: [4 1]
DEEPMD INFO _____ _____ __ __ _____ _ _ _
DEEPMD INFO | __ \ | __ \ | \/ || __ \ | | (_)| |
DEEPMD INFO | | | | ___ ___ | |__) || \ / || | | | ______ | | __ _ | |_
DEEPMD INFO | | | | / _ \ / _ \| ___/ | |\/| || | | ||______|| |/ /| || __|
DEEPMD INFO | |__| || __/| __/| | | | | || |__| | | < | || |_
DEEPMD INFO |_____/ \___| \___||_| |_| |_||_____/ |_|\_\|_| \__|
DEEPMD INFO Please read and cite:
DEEPMD INFO Wang, Zhang, Han and E, Comput.Phys.Comm. 228, 178-184 (2018)
DEEPMD INFO installed to: /home/conda/feedstock_root/build_artifacts/deepmd-kit_1678943793317/work/_skbuild/linux-x86_64-3.10/cmake-install
DEEPMD INFO source : v2.2.1
DEEPMD INFO source brach: HEAD
DEEPMD INFO source commit: 3ac8c4c7
DEEPMD INFO source commit at: 2023-03-16 12:33:24 +0800
DEEPMD INFO build float prec: double
DEEPMD INFO build variant: cuda
DEEPMD INFO build with tf inc: /opt/deepmd-kit-2.2.1/lib/python3.10/site-packages/tensorflow/include;/opt/deepmd-kit-2.2.1/lib/python3.10/site-packages/tensorflow/../../../../include
DEEPMD INFO build with tf lib:
DEEPMD INFO ---Summary of the training---------------------------------------
DEEPMD INFO running on: bohrium-14076-1013950
DEEPMD INFO computing device: cpu:0
DEEPMD INFO CUDA_VISIBLE_DEVICES: unset
DEEPMD INFO Count of visible GPU: 0
DEEPMD INFO num_intra_threads: 0
DEEPMD INFO num_inter_threads: 0
DEEPMD INFO -----------------------------------------------------------------
DEEPMD INFO ---Summary of DataSystem: training -----------------------------------------------
DEEPMD INFO found 1 system(s):
DEEPMD INFO system natoms bch_sz n_bch prob pbc
DEEPMD INFO ../00.data/training_data 5 7 23 1.000 T
DEEPMD INFO --------------------------------------------------------------------------------------
DEEPMD INFO ---Summary of DataSystem: validation -----------------------------------------------
DEEPMD INFO found 1 system(s):
DEEPMD INFO system natoms bch_sz n_bch prob pbc
DEEPMD INFO ../00.data/validation_data 5 7 5 1.000 T
DEEPMD INFO --------------------------------------------------------------------------------------
DEEPMD INFO training without frame parameter
DEEPMD INFO data stating... (this step may take long time)
OMP: Info #254: KMP_AFFINITY: pid 118 tid 118 thread 0 bound to OS proc set 0
DEEPMD INFO built lr
DEEPMD INFO built network
DEEPMD INFO built training
WARNING:root:To get the best performance, it is recommended to adjust the number of threads by setting the environment variables OMP_NUM_THREADS, TF_INTRA_OP_PARALLELISM_THREADS, and TF_INTER_OP_PARALLELISM_THREADS. See https://deepmd.rtfd.io/parallelism/ for more information.
DEEPMD INFO initialize model from scratch
DEEPMD INFO start training at lr 1.00e-03 (== 1.00e-03), decay_step 50, decay_rate 0.950006, final lr will be 3.51e-08
DEEPMD INFO batch 200 training time 6.10 s, testing time 0.02 s
DEEPMD INFO batch 400 training time 4.83 s, testing time 0.02 s
DEEPMD INFO batch 600 training time 4.84 s, testing time 0.02 s
DEEPMD INFO batch 800 training time 4.85 s, testing time 0.02 s
DEEPMD INFO batch 1000 training time 4.85 s, testing time 0.02 s
DEEPMD INFO saved checkpoint model.ckpt
DEEPMD INFO batch 1200 training time 4.86 s, testing time 0.02 s
DEEPMD INFO batch 1400 training time 5.39 s, testing time 0.02 s
DEEPMD INFO batch 1600 training time 4.84 s, testing time 0.02 s
DEEPMD INFO batch 1800 training time 4.86 s, testing time 0.02 s
DEEPMD INFO batch 2000 training time 4.84 s, testing time 0.02 s
DEEPMD INFO saved checkpoint model.ckpt
DEEPMD INFO batch 2200 training time 4.86 s, testing time 0.02 s
DEEPMD INFO batch 2400 training time 4.90 s, testing time 0.02 s
DEEPMD INFO batch 2600 training time 4.87 s, testing time 0.02 s
DEEPMD INFO batch 2800 training time 4.84 s, testing time 0.02 s
DEEPMD INFO batch 3000 training time 4.86 s, testing time 0.02 s
DEEPMD INFO saved checkpoint model.ckpt
DEEPMD INFO batch 3200 training time 4.86 s, testing time 0.02 s
DEEPMD INFO batch 3400 training time 4.99 s, testing time 0.02 s
DEEPMD INFO batch 3600 training time 4.88 s, testing time 0.02 s
DEEPMD INFO batch 3800 training time 4.85 s, testing time 0.02 s
DEEPMD INFO batch 4000 training time 4.88 s, testing time 0.02 s
DEEPMD INFO saved checkpoint model.ckpt
DEEPMD INFO batch 4200 training time 4.88 s, testing time 0.02 s
DEEPMD INFO batch 4400 training time 4.86 s, testing time 0.02 s
DEEPMD INFO batch 4600 training time 4.92 s, testing time 0.02 s
DEEPMD INFO batch 4800 training time 4.86 s, testing time 0.02 s
DEEPMD INFO batch 5000 training time 4.86 s, testing time 0.02 s
DEEPMD INFO saved checkpoint model.ckpt
DEEPMD INFO batch 5200 training time 4.87 s, testing time 0.02 s
DEEPMD INFO batch 5400 training time 4.88 s, testing time 0.02 s
DEEPMD INFO batch 5600 training time 4.87 s, testing time 0.02 s
DEEPMD INFO batch 5800 training time 4.87 s, testing time 0.02 s
DEEPMD INFO batch 6000 training time 4.90 s, testing time 0.02 s
WARNING:tensorflow:From /opt/deepmd-kit-2.2.1/lib/python3.10/site-packages/tensorflow/python/training/saver.py:1066: remove_checkpoint (from tensorflow.python.training.checkpoint_management) is deprecated and will be removed in a future version.
Instructions for updating:
Use standard file APIs to delete files with this prefix.
WARNING:tensorflow:From /opt/deepmd-kit-2.2.1/lib/python3.10/site-packages/tensorflow/python/training/saver.py:1066: remove_checkpoint (from tensorflow.python.training.checkpoint_management) is deprecated and will be removed in a future version.
Instructions for updating:
Use standard file APIs to delete files with this prefix.
DEEPMD INFO saved checkpoint model.ckpt
DEEPMD INFO batch 6200 training time 4.86 s, testing time 0.02 s
DEEPMD INFO batch 6400 training time 4.87 s, testing time 0.02 s
DEEPMD INFO batch 6600 training time 4.86 s, testing time 0.02 s
DEEPMD INFO batch 6800 training time 4.84 s, testing time 0.02 s
DEEPMD INFO batch 7000 training time 4.93 s, testing time 0.02 s
DEEPMD INFO saved checkpoint model.ckpt
DEEPMD INFO batch 7200 training time 4.89 s, testing time 0.02 s
DEEPMD INFO batch 7400 training time 4.88 s, testing time 0.02 s
DEEPMD INFO batch 7600 training time 4.88 s, testing time 0.02 s
DEEPMD INFO batch 7800 training time 4.87 s, testing time 0.02 s
DEEPMD INFO batch 8000 training time 4.86 s, testing time 0.02 s
DEEPMD INFO saved checkpoint model.ckpt
DEEPMD INFO batch 8200 training time 4.87 s, testing time 0.02 s
DEEPMD INFO batch 8400 training time 4.85 s, testing time 0.02 s
DEEPMD INFO batch 8600 training time 4.86 s, testing time 0.02 s
DEEPMD INFO batch 8800 training time 4.87 s, testing time 0.02 s
DEEPMD INFO batch 9000 training time 4.83 s, testing time 0.02 s
DEEPMD INFO saved checkpoint model.ckpt
DEEPMD INFO batch 9200 training time 4.87 s, testing time 0.02 s
DEEPMD INFO batch 9400 training time 4.95 s, testing time 0.02 s
DEEPMD INFO batch 9600 training time 4.89 s, testing time 0.02 s
DEEPMD INFO batch 9800 training time 4.87 s, testing time 0.02 s
DEEPMD INFO batch 10000 training time 4.88 s, testing time 0.02 s
DEEPMD INFO saved checkpoint model.ckpt
DEEPMD INFO average training time: 0.0244 s/batch (exclude first 200 batches)
DEEPMD INFO finished training
DEEPMD INFO wall time: 256.669 s
On the screen, you will see the information of the data system(s)
DEEPMD INFO -----------------------------------------------------------------
DEEPMD INFO ---Summary of DataSystem: training ----------------------------------
DEEPMD INFO found 1 system(s):
DEEPMD INFO system natoms bch_sz n_bch prob pbc
DEEPMD INFO ../00.data/training_data 5 7 23 1.000 T
DEEPMD INFO -------------------------------------------------------------------------
DEEPMD INFO ---Summary of DataSystem: validation ----------------------------------
DEEPMD INFO found 1 system(s):
DEEPMD INFO system natoms bch_sz n_bch prob pbc
DEEPMD INFO ../00.data/validation_data 5 7 5 1.000 T
DEEPMD INFO -------------------------------------------------------------------------
and the starting and final learning rate of this training
DEEPMD INFO start training at lr 1.00e-03 (== 1.00e-03), decay_step 50, decay_rate 0.950006, final lr will be 3.51e-08
If everything works fine, you will see, on the screen, information printed every 1000 steps, like
DEEPMD INFO batch 200 training time 6.04 s, testing time 0.02 s
DEEPMD INFO batch 400 training time 4.80 s, testing time 0.02 s
DEEPMD INFO batch 600 training time 4.80 s, testing time 0.02 s
DEEPMD INFO batch 800 training time 4.78 s, testing time 0.02 s
DEEPMD INFO batch 1000 training time 4.77 s, testing time 0.02 s
DEEPMD INFO saved checkpoint model.ckpt
DEEPMD INFO batch 1200 training time 4.47 s, testing time 0.02 s
DEEPMD INFO batch 1400 training time 4.49 s, testing time 0.02 s
DEEPMD INFO batch 1600 training time 4.45 s, testing time 0.02 s
DEEPMD INFO batch 1800 training time 4.44 s, testing time 0.02 s
DEEPMD INFO batch 2000 training time 4.46 s, testing time 0.02 s
DEEPMD INFO saved checkpoint model.ckpt
They present the training and testing time counts. At the end of the 1000th batch, the model is saved in Tensorflow’s checkpoint file model.ckpt. At the same time, the training and testing errors are presented in file lcurve.out.
The file contains 8 columns, form left to right, are the training step, the validation loss, training loss, root mean square (RMS) validation error of energy, RMS training error of energy, RMS validation error of force, RMS training error of force and the learning rate. The RMS error (RMSE) of the energy is normalized by number of atoms in the system.
head -n 2 lcurve.out
# step rmse_val rmse_trn rmse_e_val rmse_e_trn rmse_f_val rmse_f_trn lr
0 2.02e+01 1.51e+01 1.37e-01 1.41e-01 6.40e-01 4.79e-01 1.0e-03
and
$ tail -n 2 lcurve.out
9800 2.45e-02 4.02e-02 3.20e-04 3.88e-04 2.40e-02 3.94e-02 4.3e-08
10000 4.60e-02 3.76e-02 8.65e-04 5.35e-04 4.52e-02 3.69e-02 3.5e-08
Volumes 4, 5 and 6, 7 present energy and force training and testing errors, respectively.
! head -n 2 lcurve.out && tail -n 2 lcurve.out
# step rmse_val rmse_trn rmse_e_val rmse_e_trn rmse_f_val rmse_f_trn lr
0 2.06e+01 1.94e+01 1.34e-01 1.35e-01 6.51e-01 6.14e-01 1.0e-03
9800 5.49e-02 4.00e-02 7.55e-04 7.28e-04 5.37e-02 3.91e-02 4.3e-08
10000 6.56e-02 6.37e-02 1.13e-03 1.54e-03 6.44e-02 6.25e-02 3.5e-08
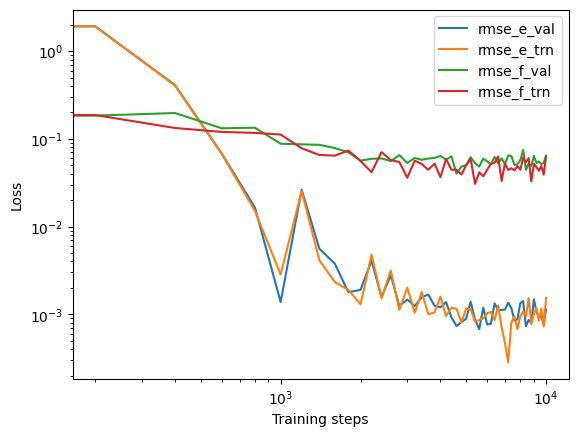
The loss function can be visualized to monitor the training process.
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
with open("lcurve.out") as f:
headers = f.readline().split()[1:]
lcurve = pd.DataFrame(np.loadtxt("lcurve.out"), columns=headers)
legends = ["rmse_e_val", "rmse_e_trn", "rmse_f_val", "rmse_f_trn"]
for legend in legends:
plt.loglog(lcurve["step"], lcurve[legend], label=legend)
plt.legend()
plt.xlabel("Training steps")
plt.ylabel("Loss")
plt.show()
2.8. Freeze a model
At the end of the training, the model parameters saved in TensorFlow’s checkpoint file should be frozen as a model file that is usually ended with extension .pb. Simply execute
! dp freeze -o graph.pb
WARNING:tensorflow:From /opt/deepmd-kit-2.2.1/lib/python3.10/site-packages/tensorflow/python/compat/v2_compat.py:107: disable_resource_variables (from tensorflow.python.ops.variable_scope) is deprecated and will be removed in a future version.
Instructions for updating:
non-resource variables are not supported in the long term
WARNING:root:To get the best performance, it is recommended to adjust the number of threads by setting the environment variables OMP_NUM_THREADS, TF_INTRA_OP_PARALLELISM_THREADS, and TF_INTER_OP_PARALLELISM_THREADS. See https://deepmd.rtfd.io/parallelism/ for more information.
WARNING:root:Environment variable KMP_BLOCKTIME is empty. Use the default value 0
WARNING:root:Environment variable KMP_AFFINITY is empty. Use the default value granularity=fine,verbose,compact,1,0
/opt/deepmd-kit-2.2.1/lib/python3.10/importlib/__init__.py:169: UserWarning: The NumPy module was reloaded (imported a second time). This can in some cases result in small but subtle issues and is discouraged.
_bootstrap._exec(spec, module)
2023-04-20 23:40:25.666203: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /usr/local/nvidia/lib:/usr/local/nvidia/lib64
2023-04-20 23:40:25.666257: W tensorflow/stream_executor/cuda/cuda_driver.cc:269] failed call to cuInit: UNKNOWN ERROR (303)
DEEPMD INFO The following nodes will be frozen: ['model_type', 'descrpt_attr/rcut', 'descrpt_attr/ntypes', 'model_attr/tmap', 'model_attr/model_type', 'model_attr/model_version', 'train_attr/min_nbor_dist', 'train_attr/training_script', 'o_energy', 'o_force', 'o_virial', 'o_atom_energy', 'o_atom_virial', 'fitting_attr/dfparam', 'fitting_attr/daparam']
WARNING:tensorflow:From /opt/deepmd-kit-2.2.1/lib/python3.10/site-packages/deepmd/entrypoints/freeze.py:354: convert_variables_to_constants (from tensorflow.python.framework.graph_util_impl) is deprecated and will be removed in a future version.
Instructions for updating:
Use `tf.compat.v1.graph_util.convert_variables_to_constants`
WARNING:tensorflow:From /opt/deepmd-kit-2.2.1/lib/python3.10/site-packages/deepmd/entrypoints/freeze.py:354: convert_variables_to_constants (from tensorflow.python.framework.graph_util_impl) is deprecated and will be removed in a future version.
Instructions for updating:
Use `tf.compat.v1.graph_util.convert_variables_to_constants`
WARNING:tensorflow:From /opt/deepmd-kit-2.2.1/lib/python3.10/site-packages/tensorflow/python/framework/convert_to_constants.py:925: extract_sub_graph (from tensorflow.python.framework.graph_util_impl) is deprecated and will be removed in a future version.
Instructions for updating:
Use `tf.compat.v1.graph_util.extract_sub_graph`
WARNING:tensorflow:From /opt/deepmd-kit-2.2.1/lib/python3.10/site-packages/tensorflow/python/framework/convert_to_constants.py:925: extract_sub_graph (from tensorflow.python.framework.graph_util_impl) is deprecated and will be removed in a future version.
Instructions for updating:
Use `tf.compat.v1.graph_util.extract_sub_graph`
DEEPMD INFO 1211 ops in the final graph.
and it will output a model file named graph.pb in the current directory.
2.9. Test a model
We can check the quality of the trained model by running
! dp test -m graph.pb -s ../00.data/validation_data
WARNING:tensorflow:From /opt/deepmd-kit-2.2.1/lib/python3.10/site-packages/tensorflow/python/compat/v2_compat.py:107: disable_resource_variables (from tensorflow.python.ops.variable_scope) is deprecated and will be removed in a future version.
Instructions for updating:
non-resource variables are not supported in the long term
WARNING:root:To get the best performance, it is recommended to adjust the number of threads by setting the environment variables OMP_NUM_THREADS, TF_INTRA_OP_PARALLELISM_THREADS, and TF_INTER_OP_PARALLELISM_THREADS. See https://deepmd.rtfd.io/parallelism/ for more information.
WARNING:root:Environment variable KMP_BLOCKTIME is empty. Use the default value 0
WARNING:root:Environment variable KMP_AFFINITY is empty. Use the default value granularity=fine,verbose,compact,1,0
/opt/deepmd-kit-2.2.1/lib/python3.10/importlib/__init__.py:169: UserWarning: The NumPy module was reloaded (imported a second time). This can in some cases result in small but subtle issues and is discouraged.
_bootstrap._exec(spec, module)
2023-04-20 23:40:30.102300: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /usr/local/nvidia/lib:/usr/local/nvidia/lib64
2023-04-20 23:40:30.102346: W tensorflow/stream_executor/cuda/cuda_driver.cc:269] failed call to cuInit: UNKNOWN ERROR (303)
WARNING:tensorflow:From /opt/deepmd-kit-2.2.1/lib/python3.10/site-packages/deepmd/utils/batch_size.py:61: is_gpu_available (from tensorflow.python.framework.test_util) is deprecated and will be removed in a future version.
Instructions for updating:
Use `tf.config.list_physical_devices('GPU')` instead.
WARNING:tensorflow:From /opt/deepmd-kit-2.2.1/lib/python3.10/site-packages/deepmd/utils/batch_size.py:61: is_gpu_available (from tensorflow.python.framework.test_util) is deprecated and will be removed in a future version.
Instructions for updating:
Use `tf.config.list_physical_devices('GPU')` instead.
DEEPMD WARNING You can use the environment variable DP_INFER_BATCH_SIZE tocontrol the inference batch size (nframes * natoms). The default value is 1024.
DEEPMD INFO # ---------------output of dp test---------------
DEEPMD INFO # testing system : ../00.data/validation_data
OMP: Info #155: KMP_AFFINITY: Initial OS proc set respected: 0-7
OMP: Info #216: KMP_AFFINITY: decoding x2APIC ids.
OMP: Info #216: KMP_AFFINITY: cpuid leaf 11 not supported.
OMP: Info #216: KMP_AFFINITY: decoding legacy APIC ids.
OMP: Info #157: KMP_AFFINITY: 8 available OS procs
OMP: Info #158: KMP_AFFINITY: Uniform topology
OMP: Info #287: KMP_AFFINITY: topology layer "LL cache" is equivalent to "socket".
OMP: Info #192: KMP_AFFINITY: 1 socket x 4 cores/socket x 2 threads/core (4 total cores)
OMP: Info #218: KMP_AFFINITY: OS proc to physical thread map:
OMP: Info #172: KMP_AFFINITY: OS proc 0 maps to socket 0 core 0 thread 0
OMP: Info #172: KMP_AFFINITY: OS proc 1 maps to socket 0 core 0 thread 1
OMP: Info #172: KMP_AFFINITY: OS proc 2 maps to socket 0 core 1 thread 0
OMP: Info #172: KMP_AFFINITY: OS proc 3 maps to socket 0 core 1 thread 1
OMP: Info #172: KMP_AFFINITY: OS proc 4 maps to socket 0 core 2 thread 0
OMP: Info #172: KMP_AFFINITY: OS proc 5 maps to socket 0 core 2 thread 1
OMP: Info #172: KMP_AFFINITY: OS proc 6 maps to socket 0 core 3 thread 0
OMP: Info #172: KMP_AFFINITY: OS proc 7 maps to socket 0 core 3 thread 1
OMP: Info #254: KMP_AFFINITY: pid 254 tid 265 thread 1 bound to OS proc set 2
OMP: Info #254: KMP_AFFINITY: pid 254 tid 267 thread 2 bound to OS proc set 4
OMP: Info #254: KMP_AFFINITY: pid 254 tid 268 thread 3 bound to OS proc set 6
OMP: Info #254: KMP_AFFINITY: pid 254 tid 269 thread 4 bound to OS proc set 1
OMP: Info #254: KMP_AFFINITY: pid 254 tid 270 thread 5 bound to OS proc set 3
OMP: Info #254: KMP_AFFINITY: pid 254 tid 271 thread 6 bound to OS proc set 5
OMP: Info #254: KMP_AFFINITY: pid 254 tid 272 thread 7 bound to OS proc set 7
OMP: Info #254: KMP_AFFINITY: pid 254 tid 273 thread 8 bound to OS proc set 0
DEEPMD INFO # number of test data : 40
DEEPMD INFO Energy MAE : 4.400922e-03 eV
DEEPMD INFO Energy RMSE : 5.258026e-03 eV
DEEPMD INFO Energy MAE/Natoms : 8.801843e-04 eV
DEEPMD INFO Energy RMSE/Natoms : 1.051605e-03 eV
DEEPMD INFO Force MAE : 4.277741e-02 eV/A
DEEPMD INFO Force RMSE : 5.514855e-02 eV/A
DEEPMD INFO Virial MAE : 6.080471e-02 eV
DEEPMD INFO Virial RMSE : 7.882116e-02 eV
DEEPMD INFO Virial MAE/Natoms : 1.216094e-02 eV
DEEPMD INFO Virial RMSE/Natoms : 1.576423e-02 eV
DEEPMD INFO # -----------------------------------------------
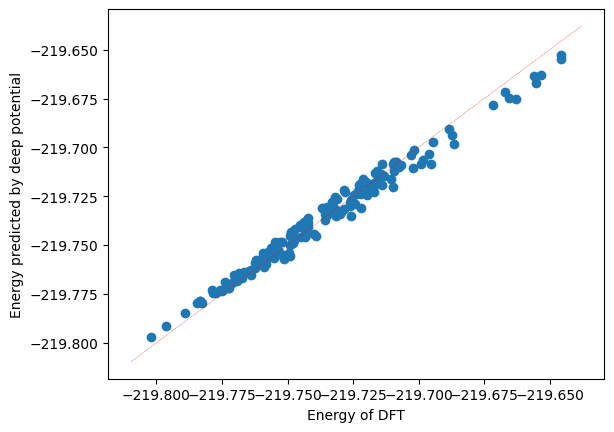
The correlation between predicted data and original data can also be calculated.
import dpdata
training_systems = dpdata.LabeledSystem("../00.data/training_data", fmt="deepmd/npy")
predict = training_systems.predict("graph.pb")
2023-04-20 23:40:32.104716: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
2023-04-20 23:40:34.426193: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer.so.7'; dlerror: libnvinfer.so.7: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /usr/local/nvidia/lib:/usr/local/nvidia/lib64
2023-04-20 23:40:34.427318: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer_plugin.so.7'; dlerror: libnvinfer_plugin.so.7: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /usr/local/nvidia/lib:/usr/local/nvidia/lib64
2023-04-20 23:40:34.427332: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Cannot dlopen some TensorRT libraries. If you would like to use Nvidia GPU with TensorRT, please make sure the missing libraries mentioned above are installed properly.
WARNING:tensorflow:From /opt/mamba/lib/python3.10/site-packages/tensorflow/python/compat/v2_compat.py:107: disable_resource_variables (from tensorflow.python.ops.variable_scope) is deprecated and will be removed in a future version.
Instructions for updating:
non-resource variables are not supported in the long term
WARNING:root:To get the best performance, it is recommended to adjust the number of threads by setting the environment variables OMP_NUM_THREADS, TF_INTRA_OP_PARALLELISM_THREADS, and TF_INTER_OP_PARALLELISM_THREADS. See https://deepmd.rtfd.io/parallelism/ for more information.
WARNING:tensorflow:From /opt/mamba/lib/python3.10/site-packages/deepmd/utils/batch_size.py:61: is_gpu_available (from tensorflow.python.framework.test_util) is deprecated and will be removed in a future version.
Instructions for updating:
Use `tf.config.list_physical_devices('GPU')` instead.
2023-04-20 23:40:36.161142: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
2023-04-20 23:40:36.165078: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /usr/local/nvidia/lib:/usr/local/nvidia/lib64
2023-04-20 23:40:36.165119: W tensorflow/compiler/xla/stream_executor/cuda/cuda_driver.cc:265] failed call to cuInit: UNKNOWN ERROR (303)
2023-04-20 23:40:36.165142: I tensorflow/compiler/xla/stream_executor/cuda/cuda_diagnostics.cc:156] kernel driver does not appear to be running on this host (bohrium-14076-1013950): /proc/driver/nvidia/version does not exist
2023-04-20 23:40:36.181810: I tensorflow/compiler/mlir/mlir_graph_optimization_pass.cc:357] MLIR V1 optimization pass is not enabled
WARNING:tensorflow:From /opt/mamba/lib/python3.10/site-packages/deepmd/utils/batch_size.py:61: is_gpu_available (from tensorflow.python.framework.test_util) is deprecated and will be removed in a future version.
Instructions for updating:
Use `tf.config.list_physical_devices('GPU')` instead.
WARNING:deepmd.utils.batch_size:You can use the environment variable DP_INFER_BATCH_SIZE tocontrol the inference batch size (nframes * natoms). The default value is 1024.
import matplotlib.pyplot as plt
import numpy as np
plt.scatter(training_systems["energies"], predict["energies"])
x_range = np.linspace(plt.xlim()[0], plt.xlim()[1])
plt.plot(x_range, x_range, "r--", linewidth=0.25)
plt.xlabel("Energy of DFT")
plt.ylabel("Energy predicted by deep potential")
plt.plot()
[]
2.10. Run MD with LAMMPS
The model can drive molecular dynamics in LAMMPS.
! cd ../02.lmp && cp ../01.train/graph.pb ./ && ls
conf.lmp graph.pb in.lammps
Here conf.lmp gives the initial configuration of a gas phase methane MD simulation, and the file in.lammps is the LAMMPS input script. One may check in.lammps and finds that it is a rather standard LAMMPS input file for a MD simulation, with only two exception lines:
pair_style deepmd graph.pb
pair_coeff * *
where the pair style deepmd is invoked and the model file graph.pb is provided, which means the atomic interaction will be computed by the DP model that is stored in the file graph.pb.
In an environment with a compatible version of LAMMPS, the deep potential molecular dynamics can be performed via
lmp -i input.lammps
! cd ../02.lmp && cp ../01.train/graph.pb ./ && lmp -i in.lammps
Warning:
This LAMMPS executable is in a conda environment, but the environment has
not been activated. Libraries may fail to load. To activate this environment
please see https://conda.io/activation.
LAMMPS (23 Jun 2022 - Update 1)
OMP_NUM_THREADS environment is not set. Defaulting to 1 thread. (src/comm.cpp:98)
using 1 OpenMP thread(s) per MPI task
Loaded 1 plugins from /opt/deepmd-kit-2.2.1/lib/deepmd_lmp
Reading data file ...
triclinic box = (0 0 0) to (10.114259 10.263124 10.216793) with tilt (0.036749877 0.13833062 -0.056322169)
1 by 1 by 1 MPI processor grid
reading atoms ...
5 atoms
read_data CPU = 0.004 seconds
DeePMD-kit WARNING: Environmental variable OMP_NUM_THREADS is not set. Tune OMP_NUM_THREADS for the best performance. See https://deepmd.rtfd.io/parallelism/ for more information.
Summary of lammps deepmd module ...
>>> Info of deepmd-kit:
installed to: /opt/deepmd-kit-2.2.1
source: v2.2.1
source branch: HEAD
source commit: 3ac8c4c7
source commit at: 2023-03-16 12:33:24 +0800
surpport model ver.:1.1
build variant: cuda
build with tf inc: /opt/deepmd-kit-2.2.1/include;/opt/deepmd-kit-2.2.1/include
build with tf lib: /opt/deepmd-kit-2.2.1/lib/libtensorflow_cc.so
set tf intra_op_parallelism_threads: 0
set tf inter_op_parallelism_threads: 0
>>> Info of lammps module:
use deepmd-kit at: /opt/deepmd-kit-2.2.1DeePMD-kit WARNING: Environmental variable OMP_NUM_THREADS is not set. Tune OMP_NUM_THREADS for the best performance. See https://deepmd.rtfd.io/parallelism/ for more information.
DeePMD-kit: Successfully load libcudart.so
2023-04-20 23:40:39.637091: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: SSE4.1 SSE4.2 AVX AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
2023-04-20 23:40:39.643206: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /usr/local/nvidia/lib:/usr/local/nvidia/lib64
2023-04-20 23:40:39.643234: W tensorflow/stream_executor/cuda/cuda_driver.cc:269] failed call to cuInit: UNKNOWN ERROR (303)
2023-04-20 23:40:39.643257: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:156] kernel driver does not appear to be running on this host (bohrium-14076-1013950): /proc/driver/nvidia/version does not exist
2023-04-20 23:40:39.645305: I tensorflow/core/common_runtime/process_util.cc:146] Creating new thread pool with default inter op setting: 2. Tune using inter_op_parallelism_threads for best performance.
2023-04-20 23:40:39.700559: I tensorflow/compiler/mlir/mlir_graph_optimization_pass.cc:354] MLIR V1 optimization pass is not enabled
>>> Info of model(s):
using 1 model(s): graph.pb
rcut in model: 6
ntypes in model: 2
CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE
Your simulation uses code contributions which should be cited:
- USER-DEEPMD package:
The log file lists these citations in BibTeX format.
CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE
Generated 0 of 1 mixed pair_coeff terms from geometric mixing rule
Neighbor list info ...
update every 10 steps, delay 0 steps, check no
max neighbors/atom: 2000, page size: 100000
master list distance cutoff = 7
ghost atom cutoff = 7
binsize = 3.5, bins = 3 3 3
1 neighbor lists, perpetual/occasional/extra = 1 0 0
(1) pair deepmd, perpetual
attributes: full, newton on
pair build: full/bin/atomonly
stencil: full/bin/3d
bin: standard
Setting up Verlet run ...
Unit style : metal
Current step : 0
Time step : 0.001
Per MPI rank memory allocation (min/avg/max) = 3.809 | 3.809 | 3.809 Mbytes
Step PotEng KinEng TotEng Temp Press Volume
0 -219.77011 0.025852029 -219.74426 50 -810.10259 1060.5429
100 -219.76784 0.023303362 -219.74454 45.070664 -605.50113 1060.5429
200 -219.77863 0.032400378 -219.74622 62.665059 -53.929107 1060.5429
300 -219.77403 0.027115352 -219.74692 52.443373 642.24342 1060.5429
400 -219.77126 0.023079501 -219.74818 44.637697 861.365 1060.5429
500 -219.786 0.034433001 -219.75156 66.596322 256.47994 1060.5429
600 -219.78295 0.029039598 -219.75391 56.165027 -527.21506 1060.5429
700 -219.777 0.020227709 -219.75677 39.122091 -696.11258 1060.5429
800 -219.78394 0.022893217 -219.76105 44.277408 -77.227892 1060.5429
900 -219.77998 0.015506508 -219.76447 29.990893 663.84491 1060.5429
1000 -219.78328 0.015178419 -219.7681 29.356341 482.8228 1060.5429
1100 -219.7903 0.018763273 -219.77154 36.28975 -273.19351 1060.5429
1200 -219.78639 0.012922048 -219.77347 24.992328 -577.90459 1060.5429
1300 -219.79131 0.015848131 -219.77546 30.65162 -129.85247 1060.5429
1400 -219.78829 0.011969602 -219.77632 23.150218 545.58517 1060.5429
1500 -219.78735 0.010610097 -219.77674 20.520821 356.36805 1060.5429
1600 -219.78834 0.011547453 -219.77679 22.333746 -386.08305 1060.5429
1700 -219.78549 0.0095297364 -219.77596 18.431312 -522.29867 1060.5429
1800 -219.78787 0.01302987 -219.77484 25.200865 120.83085 1060.5429
1900 -219.7845 0.012341623 -219.77216 23.869737 643.66442 1060.5429
2000 -219.7857 0.017597987 -219.76811 34.035987 255.57892 1060.5429
2100 -219.7853 0.023253088 -219.76205 44.973429 -465.61243 1060.5429
2200 -219.77987 0.024650089 -219.75522 47.675348 -708.62743 1060.5429
2300 -219.78134 0.030690759 -219.75065 59.358512 -221.82549 1060.5429
2400 -219.77737 0.029446857 -219.74792 56.952699 635.02431 1060.5429
2500 -219.768 0.022122766 -219.74587 42.787292 826.89652 1060.5429
2600 -219.77246 0.02691536 -219.74554 52.056572 168.88834 1060.5429
2700 -219.77746 0.031963987 -219.7455 61.821042 -497.33107 1060.5429
2800 -219.7733 0.02814671 -219.74515 54.438107 -792.71093 1060.5429
2900 -219.77498 0.029131114 -219.74585 56.342026 -685.23164 1060.5429
3000 -219.78212 0.034326288 -219.74779 66.38993 -20.441816 1060.5429
3100 -219.77222 0.02366469 -219.74856 45.769502 708.42782 1060.5429
3200 -219.77252 0.022334468 -219.75019 43.196742 753.3138 1060.5429
3300 -219.78538 0.032458098 -219.75292 62.776693 36.172647 1060.5429
3400 -219.78047 0.026131264 -219.75434 50.540064 -661.25487 1060.5429
3500 -219.77926 0.022926821 -219.75633 44.342401 -623.5037 1060.5429
3600 -219.78369 0.024854728 -219.75884 48.071137 74.821258 1060.5429
3700 -219.7768 0.016731114 -219.76006 32.359382 709.57785 1060.5429
3800 -219.77927 0.017595175 -219.76168 34.03055 543.56168 1060.5429
3900 -219.7864 0.023003584 -219.7634 44.490867 -230.55364 1060.5429
4000 -219.78098 0.017102387 -219.76388 33.077456 -677.85161 1060.5429
4100 -219.78581 0.020907229 -219.7649 40.436341 -343.9622 1060.5429
4200 -219.78717 0.021708329 -219.76546 41.985736 491.95578 1060.5429
4300 -219.78328 0.018229256 -219.76505 35.256916 680.5279 1060.5429
4400 -219.79007 0.024931071 -219.76514 48.21879 -26.785455 1060.5429
4500 -219.78331 0.019795452 -219.76352 38.286071 -624.98799 1060.5429
4600 -219.78094 0.0196038 -219.76134 37.915399 -584.8297 1060.5429
4700 -219.78608 0.027516802 -219.75857 53.219812 74.218844 1060.5429
4800 -219.77656 0.023488867 -219.75307 45.429446 827.4406 1060.5429
4900 -219.78039 0.032832529 -219.74755 63.500874 634.64896 1060.5429
5000 -219.78237 0.040761952 -219.7416 78.837046 -224.81626 1060.5429
Loop time of 12.1251 on 1 procs for 5000 steps with 5 atoms
Performance: 35.629 ns/day, 0.674 hours/ns, 412.369 timesteps/s
242.0% CPU use with 1 MPI tasks x 1 OpenMP threads
MPI task timing breakdown:
Section | min time | avg time | max time |%varavg| %total
---------------------------------------------------------------
Pair | 12.072 | 12.072 | 12.072 | 0.0 | 99.56
Neigh | 0.0066181 | 0.0066181 | 0.0066181 | 0.0 | 0.05
Comm | 0.012792 | 0.012792 | 0.012792 | 0.0 | 0.11
Output | 0.0044695 | 0.0044695 | 0.0044695 | 0.0 | 0.04
Modify | 0.022737 | 0.022737 | 0.022737 | 0.0 | 0.19
Other | | 0.006263 | | | 0.05
Nlocal: 5 ave 5 max 5 min
Histogram: 1 0 0 0 0 0 0 0 0 0
Nghost: 130 ave 130 max 130 min
Histogram: 1 0 0 0 0 0 0 0 0 0
Neighs: 0 ave 0 max 0 min
Histogram: 1 0 0 0 0 0 0 0 0 0
FullNghs: 20 ave 20 max 20 min
Histogram: 1 0 0 0 0 0 0 0 0 0
Total # of neighbors = 20
Ave neighs/atom = 4
Neighbor list builds = 500
Dangerous builds not checked
Total wall time: 0:00:13
